Oncology
Advancing Oncology research with dGH™ and Pinpoint FISH™ assays.
Unlocking Genomic Analysis for Structural Variant Detection in Cancer
The ability to detect low frequency, heterogenous structural variants within a solid or liquid tumor sample is crucial for understanding the underlying dynamics of oncogenic drivers and investigating the role played by mechanisms such as genomic instability, chemoresistance, copy number variation and DNA amplification, to name a few.
Both our single-cell, directional Genomic Hybridization (dGH™) and Pinpoint FISH™ assays are innovative and powerful approaches to visualize and map the structure of cancer genomes.
dGH™ probes and assays are strand-specific, cytogenomic tools that identify specific oncogenic events with industry-high resolution, providing unprecedented insights into the initiation, propagation, and evolution of cancers. The bioinformatic design and high signal-to-noise ratio of Pinpoint FISH probes and assays make them perfect complementary tools to dGH analysis.
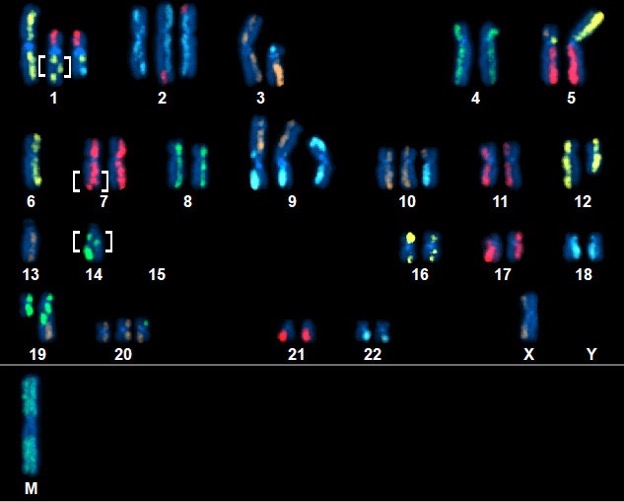
Above Fig.: Highly complex abnormal neoplastic cell featuring many abnormalities, including several small, unsuspected inversions (brackets) detected by dGH SCREEN.
Unveiling the Power of dGH Assays – Single Cell Clarity
dGH™ assays excel in their capability to map out the genomic structure of individual cells. By visualizing and enumerating genomic variants in single cells, researchers get a clearer picture of the heterogeneity present within a given cell sample instead of merely an average value. This is particularly critical in cancer research, where the structure of individual cancer cell genomes can vary widely throughout the tumor. dGH SCREEN™ (left) is composed of unique sequence, high-density (HD) dGH chromosome paints in five color panels such that chromosomes painted in the same color can be differentiated by size, shape, and centromere position. It covers all 24 chromsomes and is an ideal tool to get an unbiased view of structural variation for each single cell.
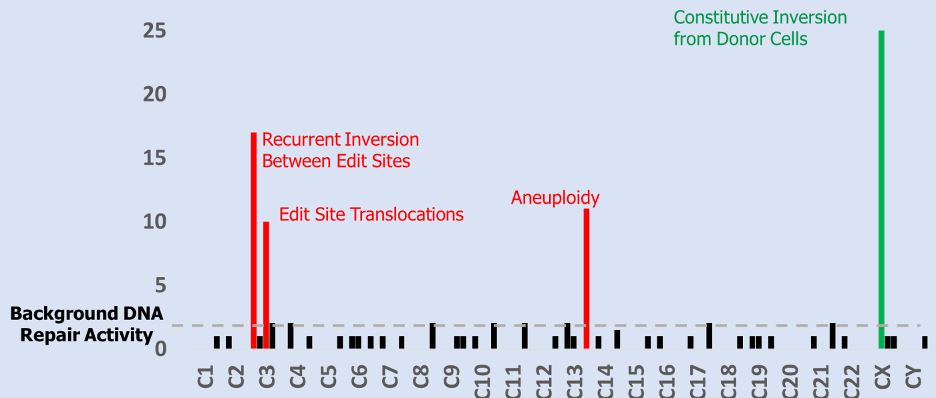
Above Fig.: Histogram displaying the numerical combination of dGH SCREEN results from many cells from one sample. Bar height indicates variant prevalence with position on the x-axis indicating relative locations along the end-to-end length of the human chromosome complement.
Detailed Analysis of Tumor Heterogeneity
By combining the genomic maps from many individual cells we provide highly detailed profiles of tumor heterogeneity, including information on genomic instability, DNA amplification and copy number variation, as well as all manner of structural variation: inversions, translocations, aneuploidy, sister chromatid exchange and other complex events. The high resolution provided by our probes and assays means that even small (<10 kb) or low frequency genomic events can be detected. KromaTiD has a team of highly skilled professionals who can provide hybridization, scoring and analysis for 100s to 1,000s of cells.
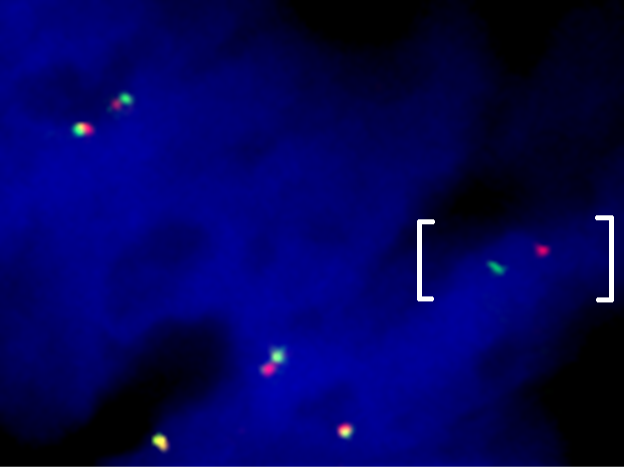
Above Fig.: Colon cancer sample hybridized with a custom break apart assay against a confidential target detecting an abnormal separation between the target loci (brackets).
Targeted Structural Variant Analysis
dGH in-Site™ assays use custom-designed or off the shelf probes targeted to identified genomic loci of interest. All KromaTiD dGH probes are synthetic and bioinformatically designed, providing low background and a high signal-to-noise ratio. In addition, the unique dGH assay cell preparation method results in parental strand-only metaphase spreads, providing the sensitivity needed to detect very small targets, down to 2 kb in some cases. These assays can be run in your own lab or through KromaTiD services.
