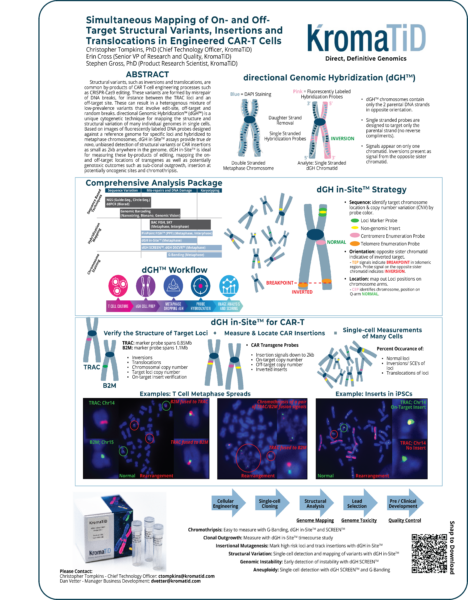
ABSTRACT
Structural variants, such as inversions and translocations, are common by-products of CAR T-cell engineering processes such as CRISPR-Cas9 editing. These variants are formed by misrepair of DNA breaks, for instance between the TRAC loci and an off-target site. These can result in a heterogenous mixture of low-prevalence variants that involve edit-site, off-target and random breaks. directional Genomic HybridizationTM (dGHTM) is a unique cytogenetic technique for mapping the structure and structural variation of many individual genomes in single cells. Based on images of fluorescently labeled DNA probes designed against a reference genome for specific loci and hybridized to metaphase chromosomes, dGH in-SiteTM assays provide true de novo, unbiased detection of structural variants or CAR insertions as small as 2kb anywhere in the genome. dGH in-SiteTM is ideal for measuring these by-products of editing, mapping the on- and off-target locations of transgenes as well as potentially genotoxic outcomes such as sub-clonal outgrowth, insertion at potentially oncogenic sites and chromothripsis.
