Strand-Specific DNA Probes – dGH Technology
Measure structural variants regardless of causation.
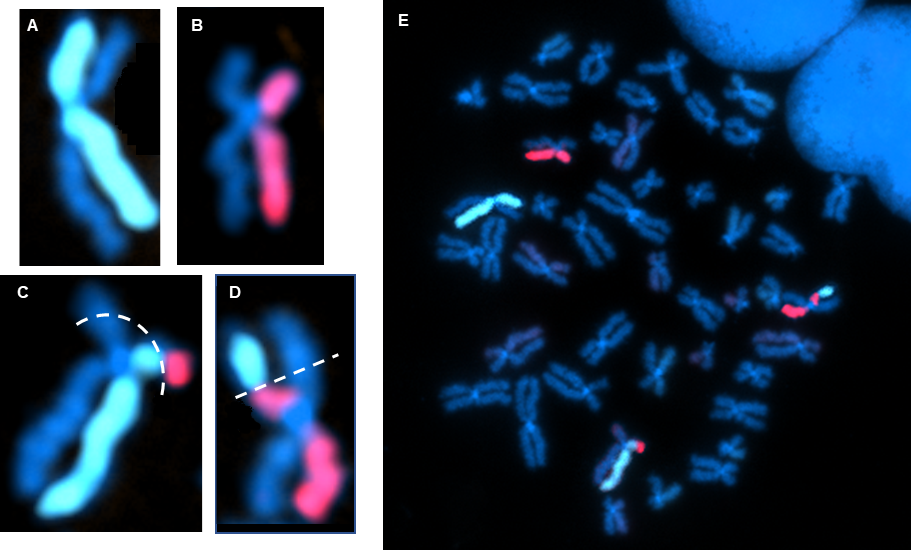
Above Fig.: dGH in-Site assay in edited CHO cells featuring custom transgene probe (green) and control probe (red). One insertion event is on an ecDNA fragment (yellow circle). The larger of the other two insertions (white circles) represents an inverted duplication.
By measuring the prevalence of random SVs on a subset of the genome, dGH can be used to determine rates of genome wide DNA damage.
directional Genomic Hybridization (dGH™) is a single-cell, cytogenetic technique where newly replicated DNA strands are selectively removed from metaphase chromosomes, resulting in single-stranded targets. When single-stranded probes are then hybridized to such targets, the resulting strand-specific hybridization can provide a level of information previously unattainable at the cytogenetic level. dGH technology can directlyvisualize all manner of genomic structural variants (SVs) including translocations, sister-chromatid exchanges, rearrangements, and previously undetectable, small inversions.
dGH Provides High-Resolution, Single-Cell Assessments of:
- Gene editing systems and their optimization
- Genomic integrity and stability
- Clonal identity
- Telomere length and aberrations
- Extra-chromosomal DNA
Inquire about a project today!
Above Fig.: For an overview of the dGH Method process: Monitoring Genomic Structural Rearrangements Resulting from Gene Editing. The dGH Method is explained in our dGH Method Application Note.
dGH is not Metaphase FISH
While the metaphase preparation is familiar to those using metaphase FISH techniques, dGH is distinguished from FISH by both the chromosome prep and the probe design. There are three primary aspects in which sample preparation differs from the traditional FISH assay. Firstly, dGH media additive is introduced to the live cell culture some hours prior to the addition of a cell cycle arresting agent. The cells will incorporate it into daughter DNA strands as they proceed through their DNA replication. Secondly, once cells are harvested and applied to slides, samples will undergo daughter-strand degradation. This involves controlled irradiation with ultraviolet light, followed by exposure to an exonuclease enzyme. Lastly, now that only parent strands remain, dGH probes targeting only one of them generate fluorescence on only that chromatid rather than on both. This generates a unique category of data traditional FISH does not provide.
Monitor Structural Variants with dGH Technology
Benefits of Direct Measurement of Structural Variants with dGH:
- Supports both unbiased and targeted genomic analysis.
- Provides complete characterization of both simple and complex structural variants regardless of sample heterogeneity.
- Single cell analysis retains the full cellular and chromosomal context.
- Direct, definitive measurement of SVs using image data without bioinformatic interpretation.
- High-resolution detection of SVs as small as 2 kb.
Key Applications for dGH Technology:
- Cell and Gene Therapy
- Biomarker Discovery and Target Validation
- Hematology / Oncology
- Biodosimetry and Environmental DNA Damage
- Rare, Undiagnosed, and Genetic Diseases
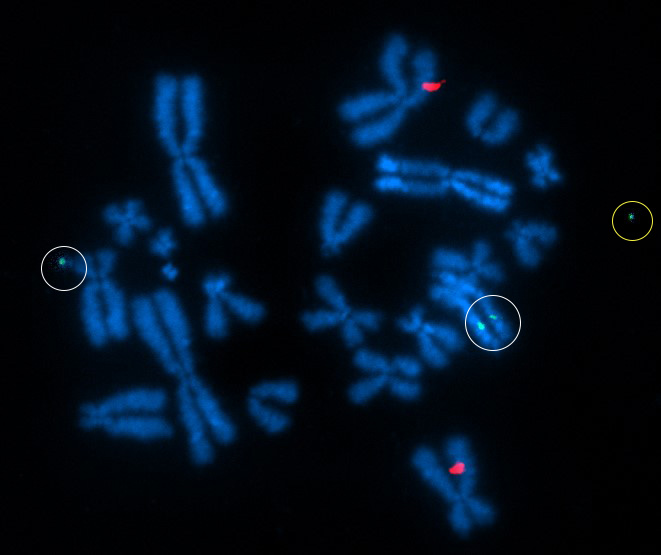
Above Fig.: Edited T cell hybridized with dGH probes for TRBC (aqua) and B2M (yellow), as well as multiple whole chromosome paints. Inversion events are indicated by a relocation of fluorescence from one chromatid to the other in one TRBC locus and in several of the painted chromosomes.
dGH and Custom Assay Solutions
dGH in-Site: Built upon the basic method of dGH, in-Site can detect and differentiate any genomic target – including inversions – as small as 2 kb. For gene therapy applications, in-Site™ is designed to track DNA cassette and transgene integration throughout the genome in an entirely de novo fashion. For edit site applications, in-Site measures any structural variation to the edit site, including rearrangements involving on-target and off-target loci.
dGH SCREEN: an unbiased dGH approach for analysis of chromosomal regions, full chromosomes, sets of chromosomes and whole genomes. Covering every unique sequence in the human genome, dGH SCREEN™ provides an un-targeted and hypothesis-free assay for the discovery, quantification and localization of any structural variant across the genome, regardless of source. dGH SCREEN is used to discover drivers of disease, measure dose effects, profile heterogenous batches of edited cells.
dGH Custom Solutions: Design genomic structural assays for analysis of specific edits or engineered cell lines. Available as a customized service for measuring any endogenous or engineered genomic target, QC of engineered cells including on- and off-target structural variations and generalized DNA damage. Telo probes can also be designed/utilized for assays involving telomeric studies.
Combining dGH with DNA stain-based assays can set structural variant data within a morphological context. With dGH technology you can examine chromosome spreads for structural abnormalities and evaluate chromosomal integrity. Stain fixed cells with DNA-specific fluorescent dyes for nuclear morphology and assessment of nuclear fragmentation. (DAPI & HOECHST)
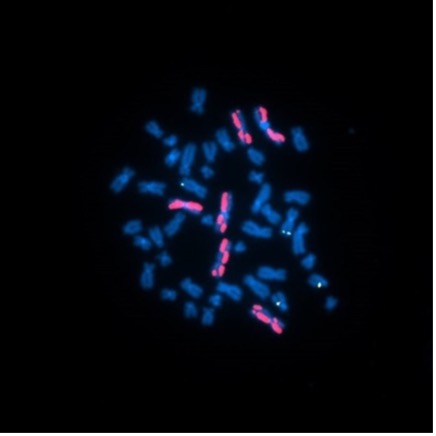
Above Fig.: dGH assay inversion detection.
Cell and Gene Therapies, Gene Editing & Cellular Engineering
Chromosomal rearrangements are a source of structural variation within the genome that are prominent in human diseases, where there is extreme importance in detecting translocations and deletions. Traditional approaches have been used for detecting these inversions but are simply not robust enough to detect. Our dGH™ technology is a single stranded hybridization method, able to detect inversions routinely, providing high resolution using our chromosome paints.
As an orthogonal analysis to PCR/sequencing techniques, dGH assays enable, through direct visualization, definitive measurement of the presence or absence of inserts. This includes genome-wide distribution and orientation of transgenes or inserted segments. When used in combination with our targeted probes and chromosome paints, dGH assays can provide quantitative, single cell on- vs. off-target outcomes of the delivery and editing process, and can be used to track the stability of the edited genome in both clonal isolates and non-clonal populations.

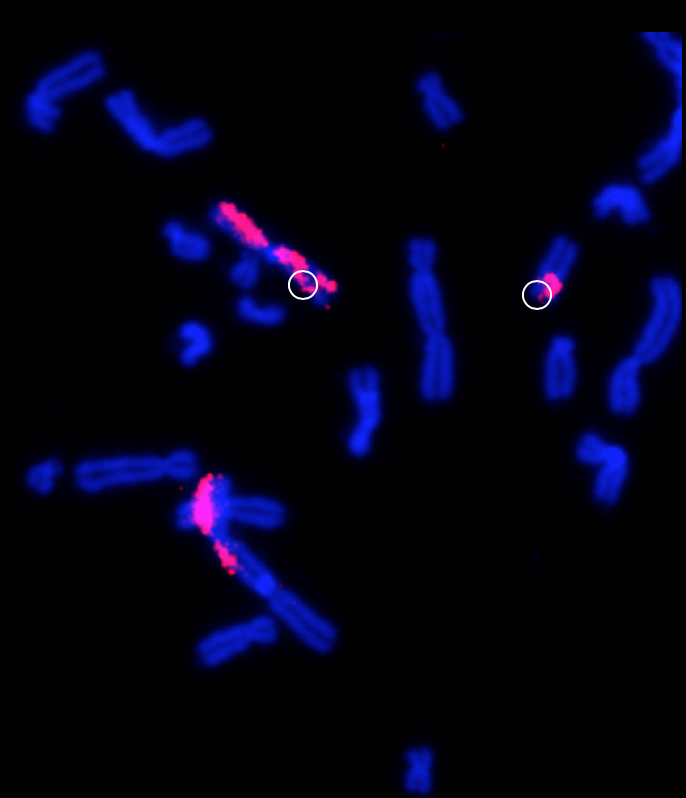
Above Fig.: Inversion detection found by dGH Technology.
Hematology & Oncology Studies
dGH assays provide single cell analysis that reliably detects low-frequency, complex variations in heterogeneous cell populations, making it ideal for identifying potentially oncogenic mutations. dGH probes generate data on all manner of structural variations, and in contrast to traditional FISH, that includes small inversions. This holds even for targets as small as 2 kb. The targets may be anywhere within the normal chromosome complement, or even on ecDNA. By targeting single-stranded sample DNA with unidirectional probes, dGH assays generate data not only on target location, but also on strand orientation and on structural changes that may otherwise go undetected by other assays. In particular at the single-digit kilobase size range, custom dGH probes can generate datasets that are otherwise out of reach, or will lack the context of their structural setting.
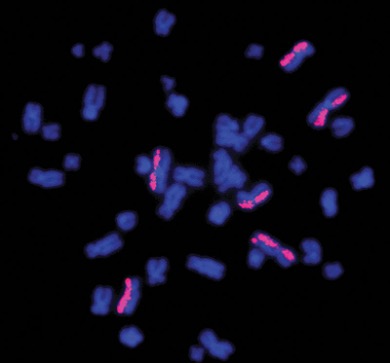
Above Fig.: Leukocyte metaphase spread hybridized with ATTO 550-labeled dGH paints. The relocation of fluorescence from one chromatid to the other in the bottom-right painted chromosome represents an inversion. This study was designed to examine the impact of ionizing radiation exposure during spaceflight on chromosomal structure in peripheral blood samples from NASA astronauts.
Biodosimetry & Environmental DNA Damage
Industrial, radioactive and environmental pollution, and public diseases can cause DNA damage. dGH is capable of providing the foundation for multiple dose response assays including inversions, dicentrics and translocations.