Cell and Gene Therapy, Gene Editing and Cellular Engineering
Robust structural variant detection and genomic integrity assays.
Careful Analysis of your Cell and Gene Therapy Research Approaches are Vital to an IND filing.
Chromosomal structural rearrangements can produce phenotypic changes which pose a risk to the recipient of a therapy product. It is vital to employ cutting-edge technologies and orthogonal data to maximize the safety profile of such products.
Single-cell dGH™ technology generates the necessary data needed for success. From unbiased, genome-wide assays, to targeted ones that can resolve targets as small as two kilobases, the dGH method allows you to directly visualize structural rearrangements, including small inversions, without any bioinformatic intermediary like sequence alignment.
dGH assays generate data orthogonal to PCR/sequencing results and enable measurement of the presence or absence of inserts through direct visualization. This includes genome-wide distribution and orientation of transgenes or inserted segments. dGH assays provide quantitative data of on- vs. off-target insertion events based on single-cell analysis.
KromaTiD’s suite of specialized assays can be used to track the stability of the edited genome in both clonal isolates and non-clonal populations. This permits an informed evaluation of your delivery and editing process.
dGH in-Site™ Targeted Assays Provide Whole Genome Tracking of DNA Cassette Inserts as Small as 2kb.
Unidirectional dGH probes provide a single cell, genome-wide perspective of cellular engineering outcomes, including tracking of viral and non-viral mediated insert integration (CRISPR/Cas and alternative systems).
- On- and off-target integration metrics
- Detects structural variants in off target CRISPR therapies
- Clean integration data, even from complex or heterogeneous cell populations
- A unique, whole genome, orthogonal method of direct visualization of inserts, without bioinformatic prediction of outcomes
- Multi-channel fluorescence for flexible and multiplex assay design
- Available for human, murine, canine, non-human primate and CHO cells.
Offering the lowest limit of detection of integrated or genomic DNA targets by fluorescence, dGH in-Site™ is the most comprehensive tool available for researchers interested in tracking engineering outcomes in a de novo fashion.
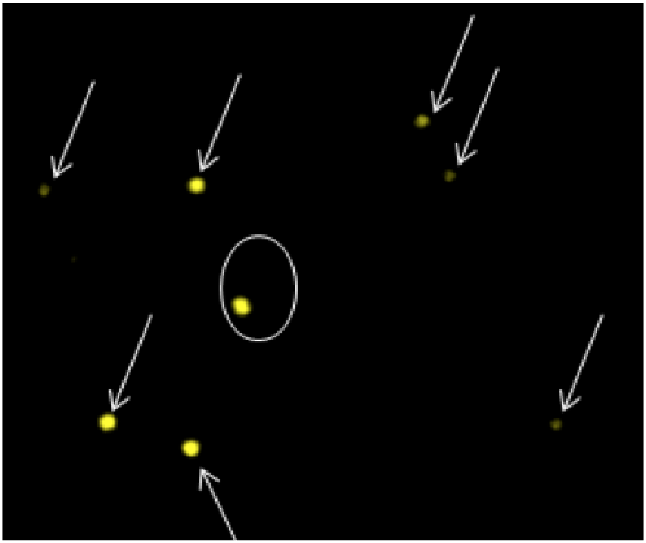
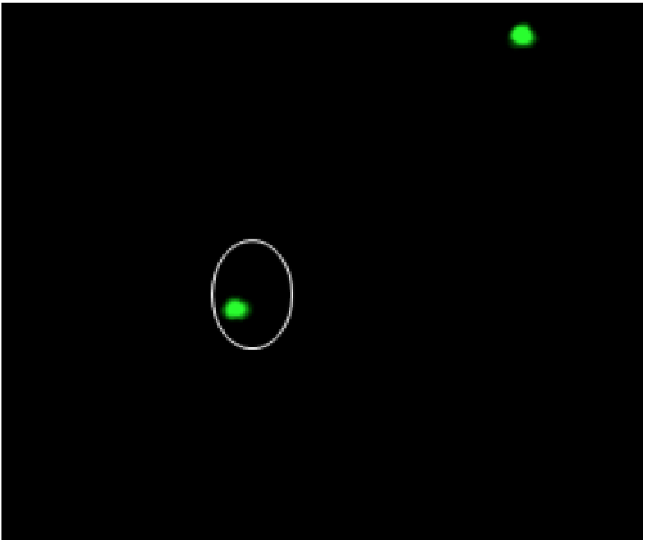
dGH in-Site™ assay in an edited iPSC, demonstrating both on-target and random integration of insert sequence (yellow) throughout the genome.
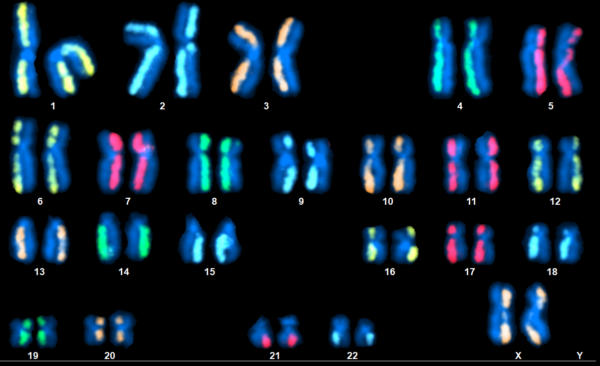
dGH SCREEN™ Unbiased Assay Designed to Measure Structural Variants throughout the Genome
dGH SCREEN™ enables direct visualization of chromosomal structural rearrangements on a single-cell basis. Rearrangements like deletions, translocations, and even small inversions are detectable without the need for any foreknowledge of likely breakpoints. This makes dGH SCREEN™ a truly unbiased tool for biomarker discovery, and direct measurement of misrepair events and broader genomic instability.
Application Benefits of SCREEN™:
- Detection of inversions and sister-chromatid exchanges
- Measurement of chromosomal Gain & Loss
- Monitoring Cellular Engineering Outcomes
- Orthogonal Data for Sequencing
- Variant Discovery
- Genomic Stability Assessment
The generous amount of numerical and structural variant data produced by dGH SCREEN™ enables the creation of variance profiles for complex and heterogeneous cell populations.
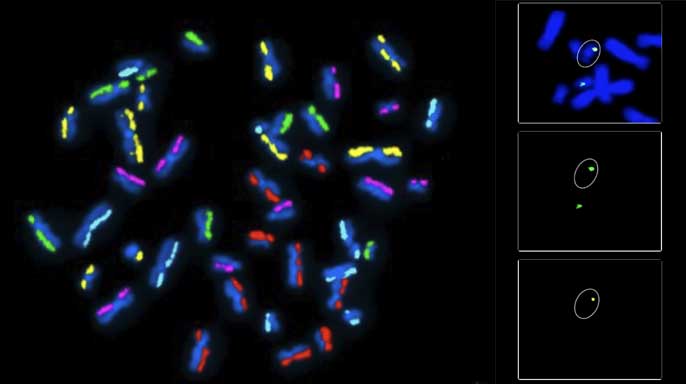
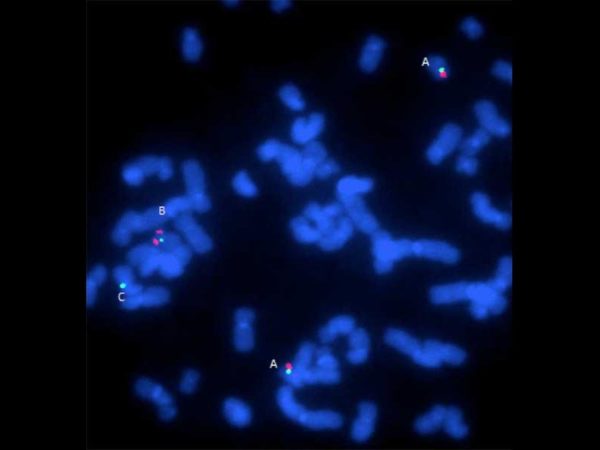
Example of the first step in the dGH SCREEN analysis process. A metaphase spread is hybridized with 5 color dGH probes designed to measure known and unknown variants in order to provide a whole genome survey of structural rearrangements in a single cell.
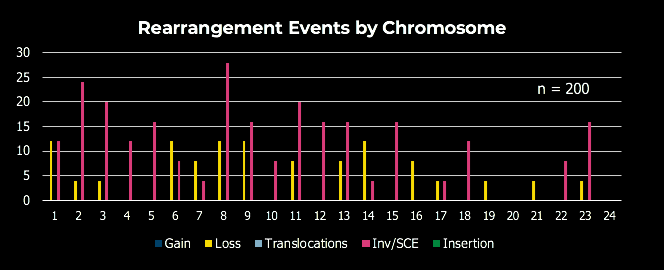
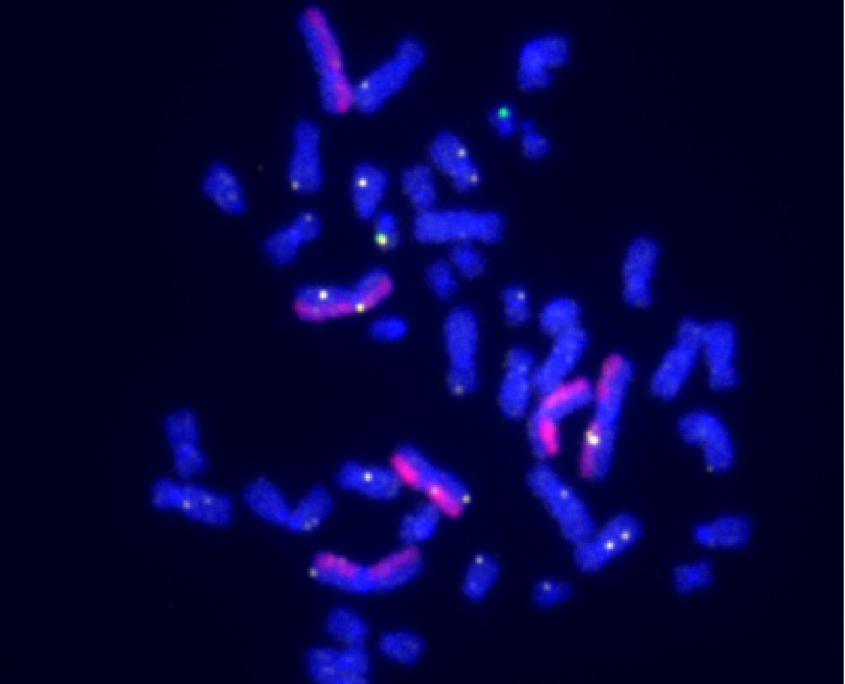
dGH SCREEN analysis example data. Chromosomes are organized karyographically and aggregate sample rearrangement data is gathered for analysis of genomic stability and structural integrity.
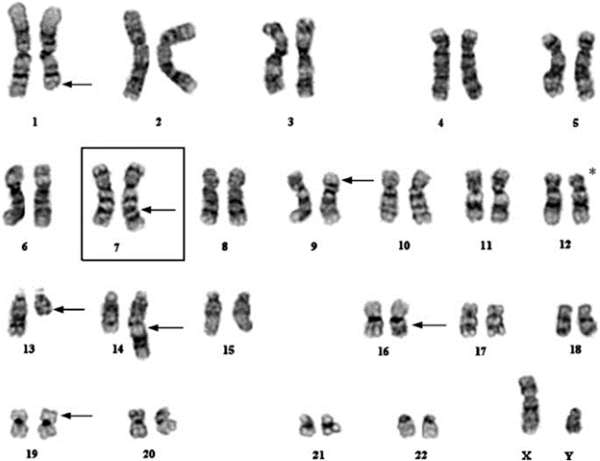
Use Genomic Integrity Karyotyping as a QC Method to Support your Cell Therapy Project.
By high-count comparison of experimental samples against a wild-type baseline, valuable genomic integrity QC data can help you stratify potential sources of genomic instability in your process. Use G-banded karyogram analysis of 100+ cells per sample to generate supporting data for your Investigational New Drug (IND) filing.
- Pivotal batch analysis
- Comprehensive reports for each individual sample
- Genomic integrity reports featuring side-by-side comparisons of your experimental samples and a wild-type baseline