dGH in-Site™ Assay Services
Track your genome editing outcomes and get a comprehensive analysis of your on- and off-target effects.
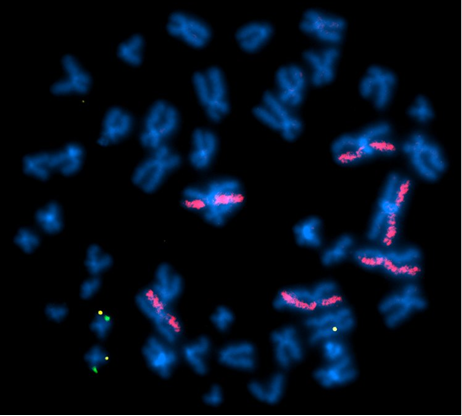
Above Fig.: Identification of On- and Off-Target Transgene Insertions: In this study, a green probe was designed to identify the intended insertion site, while a yellow probe identified the actual transgene insertion events. dGH paints (pink) generated data on the rate of structural rearrangements occurring in the host cells. This image shows on-target integration (yellow) in both copies of the AAVS1 locus on chromosome 19 (green) in addition to an off-target integration event in an unknown chromosome.
Detect and Track Any Genomic Target Starting at 2 kb.
dGH in-Site™ can target unique DNA sequences in any genomic locus, detecting challenging structural changes like small inversions in targets as small as 2 kilobases. The assay can be used to generate high-resolution, single-cell visual data on transgene integration events throughout the genome.
Our team of experts works with you to customize a project plan and probe designs to meet the genomic structural research needs unique to your project.
dGH in-Site™ Detects:
- Transgene insertions and deletions.
- Small inversions and translocations.
- Structural variants caused by mis-repair of edit sites.
- On- and off-target inserts to determine Insertional Copy Number (ICN).
Use dGH in-Site to Assess:
- Transgene integration distribution on both a single cell and genome-wide basis.
- Efficacy of standard and novel gene editing systems.
- Known disease-related DNA targets.
- Sequencing data outcomes.
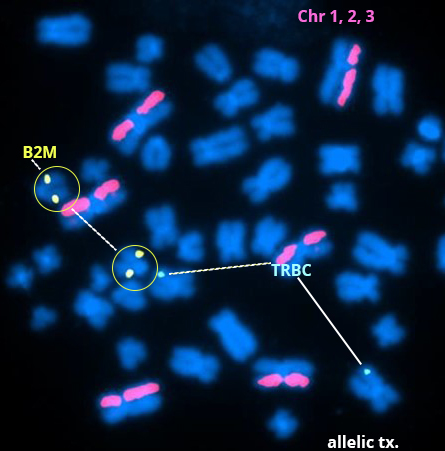
Above Fig.: Metaphase spread in edited CAR-T cells with an allelic translocation (tx.) at the B2M edit site.
Evaluate Cas-CLOVER Nuclease Allogeneic CAR T Engineering Outcomes.
Custom dGH in-Site probes targeting the TRBC edit site on chromosome 7 (cyan/aqua), and the B2M edit site on chromosome 15 (yellow) were used to evaluate the outcome of CAR-T engineering experiments using an RNA-guided endonuclease, Cas-CLOVER.
In addition to edit site specific probes, dGH chromosome paints for chromosomes 1, 2 and 3 (pink/magenta) were used to track genomic integrity. The example metaphase spread in edited CAR-T cells shown here identifies an allelic translocation (tx.) at the B2M edit site. However, overall there was an insignificant trend for increased translocation between each edit site and no significant increase in other abnormalities such as inversions, sister-chromatid exchange, deletions, gains, or insertions.
Download our Datasheet for a List of Services
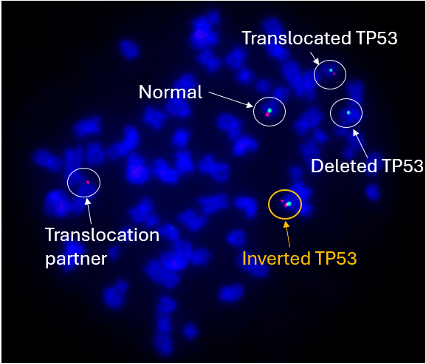
Above Fig.: dGH in-Site assay in edited TP53/CEP17 transgene probe (green) and control probe (red).
Measure and Monitor CRIPSR/Cas9 Off-Target Effects.
In order to measure the unintended effects of CRISPR/Cas9 editing process, a customized TP53/CEP17 assay using dGH in-Site probes was designed to detect edits made near the TP53 gene in HEK cells. Double-strand breaks caused by CRISPR/Cas9 can lead to deleterious structural rearrangements.
The TP53 probe (red), with the CEP17 (green) as a reference signal, detected not just the deletions and translocations a comparable FISH assay would, but also a small inversion. Metaphase FISH would only detect this change with a dedicated break apart design, and then only if the inversion was large enough to separate the signals. Depending on the target characteristics, dGH probes can be designed against loci as small as 2 kilobases, resulting in a high-resolution assay capable of detecting most classes of structural variations in a single test.
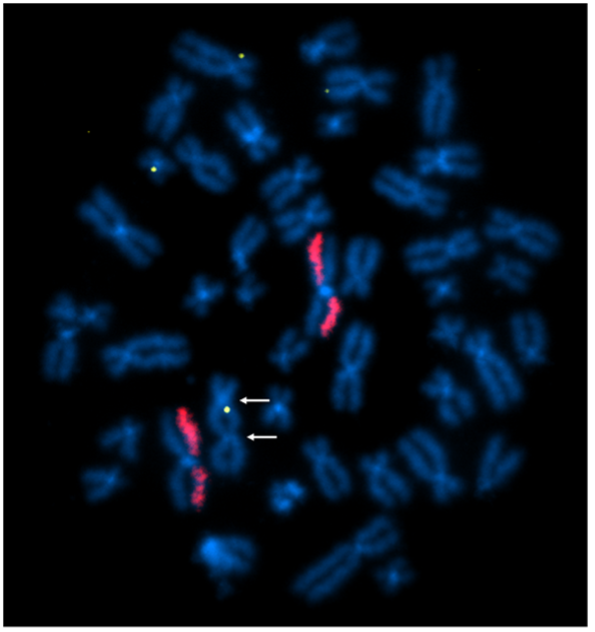
Above Fig.: In this image, four insert sequence signals are present. While the painted chromosomes show a normal signal pattern, one of the chromosomes with an insertion is a dicentric (contains two centromeres, see arrows), a structural rearrangement classically used as a metric for genotoxicity.
Genotoxic Effects of CAR-T Engineering with Lentivirus.
A custom dGH in-Site™ assay was designed to measure transgene insertion outcomes and potential genotoxic effects of the editing system using a transgene probe (yellow) and whole-chromosome paints (red). This assay simultaneously generated data on the location and quantity of insertion events, and also on structural rearrangements.
The dGH in-Site™ assay accomplished this in a few ways. First, the insert probe reported rearrangements including translocations, duplications and inversions impacting its target. Second, the whole-chromosome paints (red) in the assay generated overall genomic instability data, as indicated by the prevalence of rearrangements impacting their target chromosomes. Last, the metaphase nature of the sample itself helps reveal rearrangements, such as the dicentric chromosome seen in the image.
